Premium Only Content
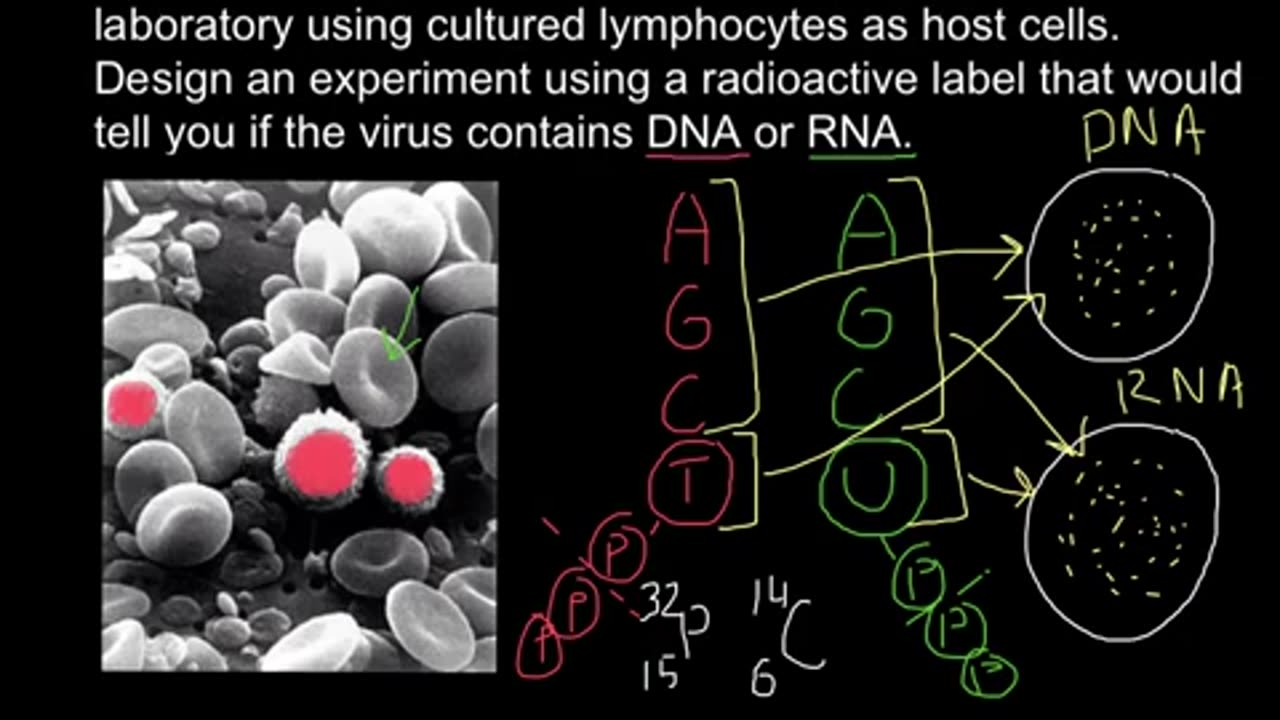
How to determine if virus DNA or RNA based using radioactive labeling?
Nucleic acids may be modified with tags that enable detection or purification. The resulting nucleic acid probes can be used to identify or recover other interacting molecules. Common labels used to generate nucleic acid probes include radioactive phosphates, biotin, fluorophores and enzymes. In addition, the bioconjugation methods used for nucleic acid probe generation may be adapted for attaching nucleic acids to other molecules or surfaces to facilitate targeted delivery or immobilization, respectively.
Overview
Enzymatic methods for nucleic acid probe labeling
Terminal deoxynucleotidyl transferase (TdT)
T4 RNA ligase
T4 polynucleotide kinase (PNK)
DNA polymerase
RNA polymerase
Chemical methods for nucleic acid probe labeling
Periodate oxidation of RNA
EDC activation of 5′ phosphate
Chemical random-labeling
Protein Methods Library Home
Overview of Protein-Nucleic Acid Interactions
Overview
Nucleic acid probes can be labeled with tags or other modifications during synthesis. However, purchasing custom oligonucleotide probes (especially RNA) can be quite expensive depending on the modification and if costly purification services are required. Additionally, the minimum order quantity for modified oligonucleotides is typically much higher than unmodified versions and may be excessive compared to the amount required for the intended application. Because of this, many researchers may choose in-house methods or labeling kits for probe generation.
Numerous reagents are available for quick and efficient bench top oligonucleotide labeling, and are most useful for making small amounts of probe or when many different probes with the same label are required (i.e., for mutational analysis). For small-scale probe generation needs, enzymatic methods are an economical method for labeling probes. In contrast, chemical methods are amenable to larger scale reactions. There are enzymatic and chemical methods for creating probes labeled at either the 5′ or 3′ ends of the oligonucleotide as well as randomly incorporated throughout the sequence. The choice of which method needed is determined in part by the degree of labeling required and if the modification will cause steric hindrance that prevents the desired interactions. Typically, nucleic acids hybridization reactions (i.e., Northern blotting) benefit from the high specific activity gained through random incorporation of label into a probe. However, assays requiring protein interactions (i.e., gel shift and pull-down assays) require end-labeling to allow protein binding.
-
 1:08:07
1:08:07
Bek Lover Podcast
2 hours agoInteresting Times with Bek Lover Podcast
5.29K -
 1:51:12
1:51:12
Tate Speech by Andrew Tate
6 hours agoEMERGENCY MEETING EPISODE 105 - UNBURDENED
130K65 -
 1:01:18
1:01:18
Tactical Advisor
8 hours agoBuilding a 308 AR10 Live! | Vault Room Live Stream 016
86.2K7 -
 2:17:02
2:17:02
Tundra Tactical
1 day ago $3.67 earnedTundra Nation Live : Shawn Of S2 Armament Joins The Boys
216K29 -
 23:22
23:22
MYLUNCHBREAK CHANNEL PAGE
2 days agoUnder The Necropolis - Pt 5
173K67 -
 54:05
54:05
TheGetCanceledPodcast
1 day ago $17.79 earnedThe GCP Ep.11 | Smack White Talks Smack DVD Vs WorldStar, Battle Rap, Universal Hood Pass & More...
174K36 -
 8:30
8:30
Game On!
13 hours ago $0.09 earnedLakers BLOCKBUSTER trade! Luka Doncic is coming to LA!
13.2K2 -
 48:29
48:29
hickok45
17 hours agoSunday Shoot-a-Round # 266
12.6K9 -
 15:18
15:18
SternAmerican
3 days agoStern American with Sam Anthony from YourNews.com
8.23K -
 1:03:13
1:03:13
PMG
1 day ago $0.12 earnedRFK, Tulsi & Kash Hearings, the Plane Crash in the Potomic, & Ozempic
7.99K2